YaliFunTome
Yarrowia lipolytica Functional Screens of Tanscription Factor-ome Database
YALI number: YALI0E07942g
Number: TF042 | Associated name: MIG1 | NCBI: 2912469 | RefSeq DNA: XM_503678.1 | RefSeq Peptide: XP_503678.1 | UniProtKB: Q6C6N4
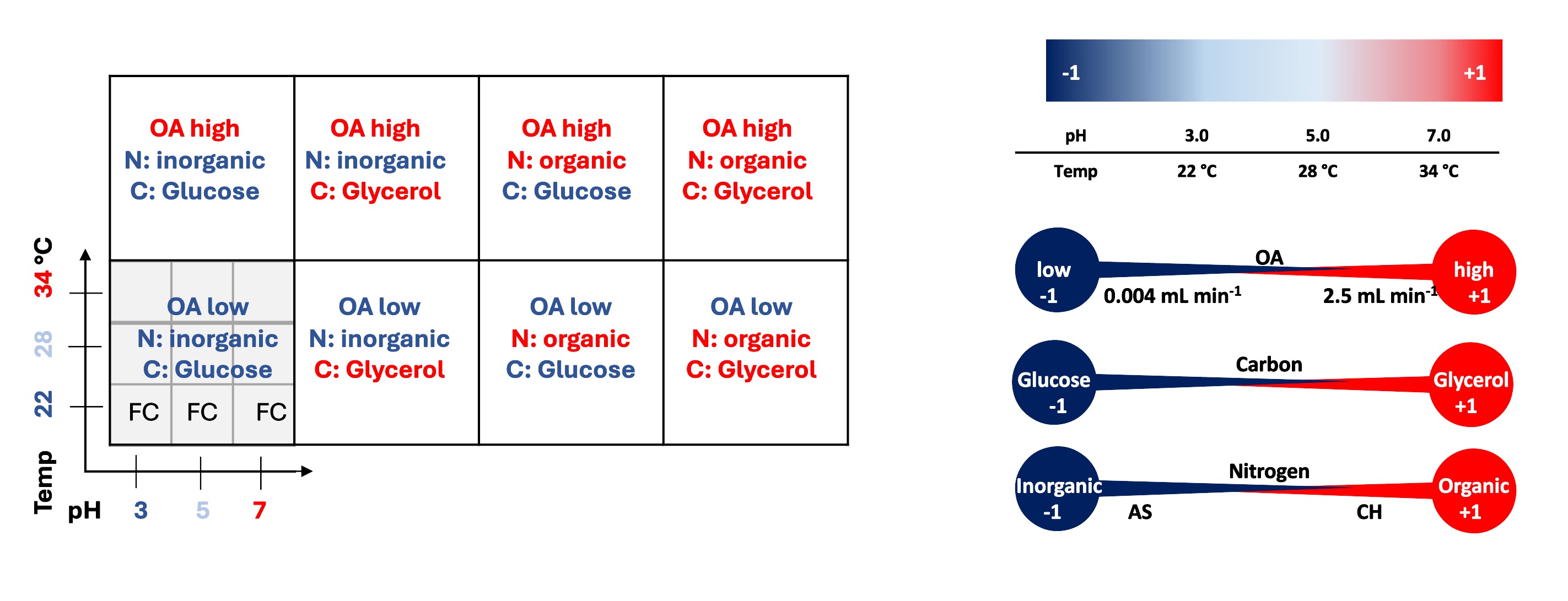
Fig.1. Graphical presentation of the arrangement of the variables' combinations corresponding to the data presentation scheme (left), and the variables' coding system (right). CH - casamino acid hydrolysate, AS - ammonium sulfate, OA- oxygen availability, FC - fold change of the response from the TF-OE strain over the control strain.
Legend:
FC - fold change over control strain, Sig - significant at p-value > 0.05, glu - glucose, gly - glycerol, i - inorganic, o - organic
| 2.0 | 1.7 | 1.5 | 1.3 | 1.1 | 1.0 | 0.7 | 0.5 | 0.3 | 0.1 |
Growth
Fold change in growth (optical density at 600 nm) of the TF-OE strain over the control strain
| 0.95 | ||
| 1 |
| 0.92 | 1.01 | |
| 0.93 | 1.02 | |
| 0.83 | ||
| 0.96 | ||
| 1.42 |
| 0.87 |
| 0.97 | ||
Factor's Contribution
| YALI0E07942g |
|---|
| OA 3.64 |
| pH 3.58 |
| Temp 1.46 |
| Carbon 0.17 |
| Nitrogen non-significant |
Total r-Prot
Fold change in fluorescence of r-Prot in the TF-OE strain over the control strain
| 1.18 | ||
| 1.05 |
| 0.97 | 1.15 | |
| 0.97 | 1.39 | |
| 0.74 | ||
| 1.01 | ||
| 1.14 |
| 0.88 |
| 0.88 | ||
Factor's Contribution
| YALI0E07942g |
|---|
| pH 8.21 |
| Carbon -0.38 |
| Temp -3.62 |
| OA -7.14 |
| Nitrogen non-significant |
Normalized r-Prot
Fold change in fluorescence of r-Prot normalized per biomass in the TF-OE strain over the control strain
| 1.1 | ||
| 1.21 |
| 1.14 | ||
| 1.06 | 0.83 |
| 0.97 | ||
| 0.97 | ||
| 0.97 |
| 0.89 | ||
| 0.98 |
| 0.94 | ||
Factor's Contribution
| YALI0E07942g |
|---|
| Temp 3.21 |
| pH -0.08 |
| Nitrogen -2.09 |
| Carbon non-significant |
| OA non-significant |